Up
Previous
Next
Day 5 Afternoon Lecture Notes
Steve Williams, Smith College
June 10, 2004
Different RevT genes from the family have sequences that are about
90% the same. When genes are copied from one part of the genome to
another, they start to diverge evolutionarily. Most copies of RevT
are non-functional "pseudogenes."
RevT codes for reverse transcriptase, which copies RevT to other
places in the genome. RevT's tendency to copy itself explains why
LINE1 has so many occurrences in the genome. Retroviruses may well be
RevT genes that escaped from the host genome. "RevT is something like
an endogenous retrovirus." If LINE1 by chance inserts into genes, it
may cause a mutation in protein production.
Primer Design Considerations
- Desirable length is 18-30 nt. Shorter primers are not specific
enough; longer primers don't work any better.
- Base composition should be near random as a primer too rich in one
base is likely not to hybridize specifically enough.
- Pick primers that don't form hairpins. These can be excluded
using a computer program. A GC-rich hairpin is more stable (and
worse) than an AT-rich one. For example
GCTACGGGGCCCCTGTG
would likely fold in the center forming four G-C bonds and fail to
hybridize with template DNA.
- Avoid primer-primer hybridization. Hairpins are an example of
intraprimer hybridization. Another example is when reverse and
forward primers bind to each other.
- Melting temperatures of the primers should be close. Primers
should be hybridized under fairly stringent conditions at
(Tm - 10°C) <= T <= (Tm - 5°C). Too
low a temperature may cause mishybridization.
- Some mismatch of the primer and template can be tolerated but not
at the 3' end. The 3' end is where synthesis and elongation start, so
if it is dangling, these processes will fail.
Forensic investigations use standard primers that hybridize to
highly conserved DNA regions but which, taken as a pair, span highly
variable regions. Some primer and software vendors: Molecular Biology
Insights (including the Oligo program), DNAsis, LaserGene, IDT
(Smith's provider) and the Primer3 program from MIT.
How to Confirm the Identity of PCR Products
- A band on a gel at the right mass is an indication of the right
product. For a routine process using a gel to check the mass is
adequate. For a new PCR process with new primers, more verification
is called for.
- Use an internal hybridization probe.
Design a 3rd nucleotide that hybridizes to the known sequence of
the expected PCR product. Often this test is performed using a
Southern blot: running a DNA gel, then transferring DNA to a membrane
where hybridization with the probe is checked.
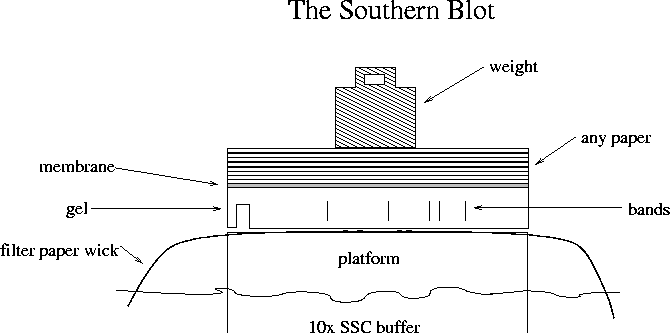
The buffer goes up the wick, through the gel, through the membrane
and into the paper. DNA follows the buffer through the gel and onto
the membrane, which it can't permeate. In so doing it moves purely
vertically and stays registered with its position in the gel. The DNA
transfer is always performed with the gel upside-down as the top
surface is too wavy to make good contact to the membrane.
The DNA on the membrane can be UV-crosslinked on for attachment and
then hybridized with a probe. The probe can be labeled with
fluorescein or biotin. Any DNA in marker lanes will also be
transferred to the membrane and will serve as a negative control for
the hybridization. Oligos can be ordered with attached probes for
added convenience. Any ethidium bromide left in the gel doesn't
interfere with the hybridization. Hybridization of a probe to a
Southern membrane should used high stringency since the method is
intended to be a strict confirmation of earlier results.
- Perform a second PCR reaction with different primers from the
first one. A second PCR reaction is a better test than Southern blot
as it requires the matching of two new oligos. This method is known
as "nested" PCR.
- Perform a restriction enzyme digest. If the PCR product is the
desired one, the products on the gel should appear at predictable
masses. The restriction enzyme method used to be known as the "DNA
fingerprint" although it has nothing to do with the forensic method
now known by that name.
- Sequence the DNA of the PCR product. Sequencing is now the method
of choice now that it's easy and cheap.
Up
Previous
Next
